CanGraph package
In this page
CanGraph package#
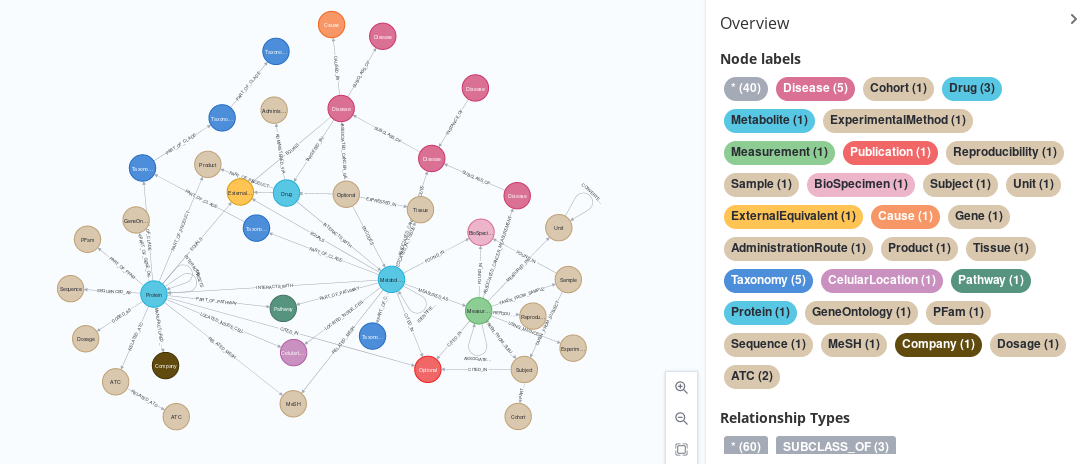
This Git Project, created as part of my Master’s Intenship at IARC, contains a series of scripts that pulls information from a series of five databases from their native format (XML, CSV, etc) into a common, GraphML format, using a shared schema that has been defined to minimize the number of repeated nodes and properties. This databases are:
Exposome-Explorer: A hand-curated, high-quality database of associations between metabolites, food intakes and outakes and different diseases, specially cancers.
Human Metabolome DataBase: An detailed, electronic database containing detailed information about small molecule metabolites found in the human body.
DrugBank: A unique bioinformatics and cheminformatics resource that combines detailed drug data with comprehensive drug target information.
Small Molecule Pathway Database: An interactive database containing more than 618 small molecule pathways found in humans, More than 70% of which are unique to this DB
WikiData: The world’s largest collaboratively generated collection of Open Data worldwide.
Each of then have their unique advantages and disadvantages (size, quality, etc) but they have been chosen to work together and help in identifying metabolites and their potential cancer associations at IARC.
With regards to the schema, it can be consulted in detail in the new-schema.graphml file, which can itself be opened in Neo4J by calling: CALL apoc.import.graphml("new-schema.graphml", {useTypes:true, storeNodeIds:false, readLabels:True}) after placing it in your Neo4J’s import directory (you can find it in the settings shown after starting the server with sudo neo4j start). It consists of a simplification of all the nodes present on the old-schema.graphml file (which itself represents the five different schemas that our five databases natively presented), arrived at by merging nodes and changing relationship names so that they are unique (and, thus, more actionable). One property, LabelName has been added as a dummy name to generate the image you can see in the header.
This repo contains two kind of scripts: first, some build_database.py scripts, which contain the information to re-build the databases in the common format from scratch, and are located in subsequent subfolders named after the database they come from (more info can be consulted on them on their respective READMEs) and a common main.py script, which can be used to query for sub-networks based solely on info presented on a sample_input.csv database of identified compounds which we would like to annotate.
Intallation#
To use this script, you should first clone it into your personal computer. The easiest way to do this is to git clone the repo:
Install git (if not already installed) and other requirements. On linux:
sudo apt install git curlClone the repo:
git clone https://codeberg.org/FlyingFlamingo/graphify-databasesStep into the directory
cd graphify-databases
Once the project has been installed, you must run setup.py, a preparation script that guides you through the process of installing all five databases on your computer, so that then we can correctly process them and generate the sub-networks. You should also install the required python modules and run the setup script:
PIP install all dependencies:
pip install -r requirements.txtRun the setup script:
python3 setup.py
Once this has been done, you are ready to start using the main script!
NOTE: If you do not wish to use git, you can manually download the repo by clicking here
Usage#
To generate this sub-networks (the original idea of the project) you should run:
python3 main.py neo4jadress databaseusername databasepassword databasefolder inputfile
where:
neo4jadress: is the URL of the database, in neo4j:// or bolt:// format
username: the username for your neo4j instance. Remember, the default is neo4j
password: the password for your database. Since the arguments are passed by BaSH onto python3, you might need to escape special characters
databasefolder: The folder indicated to
setup.pyas the one where your databases will be storedinputfile: The location of the CSV file in which the program will search for metabolites. This file should be a Comma-Separated file, with the following format:
MonoisotopicMass, SMILES, InChIKey, Name, InChI, Identifier, ChEBI
All images in this repository are CC-BY-SA-4.0 International Licensed.
NOTE: When committing to the repo, try to use GitMojis to illustrate your commit :p
Important Notices#
Some databases are auto-integrated based on their URLs. This URLs, as well as those of existing dependencies, may change over time. Please make sure to have them updated in case you want to run the latest version of the databases
We have made our best efforts to make the script as multi-platform as possible; however, the script has been developed with Linux in mind, and you may need to install additional packages if you want to run it on Windows or MacOS. Please, check the
dependenciessection for more info
Dependencies#
This python package has the following known dependencies:
Package |
Description |
Order to install |
|---|---|---|
Python 3.8 |
the python programming language |
|
cURL |
command line tool for transferring data from URLs |
|
neo4j |
a graph dbms |
|
Alternatively, as a one-liner: sudo apt install python3 curl; python3 -c'import setup; setup.setup_neo4j("neo4j", True)
Subpackages#
CanGraph.deploy module#
A python module that simplifies deploying the different formats the program can present itself as:
A web documentation that is made using sphinx
A PDF manual likewise made, also using LaTeX
A git repo where the program can be accessed and version-tagged
And, in the future… a singularity container!
CanGraph.deploy Usage#
To use this module:
usage: python3 deploy.py [-h] [-m] [-d] [-w] [-p]
Named Arguments#
- -m, --main
deploy code to your dev branch
- -d, --dev
deploy code to your main branch
- -w, --web
deploys the documentation to the ‘pages’ web site, depending on which is activated
- -p, --pdf
generates a PDF version of the sphinx manual, and saves it to the repo
Note
For this program to work, the Git environment has to be set up first.
You can ensure this by using: CanGraph.setup.setup_git
CanGraph.deploy Functions#
This module is comprised of:
- args_parser()[source]#
Parses the command line arguments into a more usable form, providing help and more
- Returns
A dictionary of the different possible options for the program as keys, specifying their set value. If no command-line arguments are provided, the help message is shown and the program exits.
- Return type
Note
Note that, in Google Docstrings, if you want a multi-line
Returnscomment, you have to start it in a different line :(Note
The return must be of type
argparse.ArgumentParserfor theargparsedirective to work and auto-gen docs
- deploy_code(branch='dev')[source]#
Deploys code from a given branch to the corresponding remote.
- Parameters
branch (str) – The name of the branch of the local git repo that we want to deploy
Note
Normally, the
pagesbranch should be published usingdeploy_webdocs, which in theory would be weird to publish without updating the docs first
- deploy_pdf_manual(docs_folder='./docs/', work_dir='.', manual_location='./CanGraph_Manual.pdf', prechecks_done=False, custom_domain=None)[source]#
Parses the command line arguments into a more usable form, providing help and more
Generates the PDF docs guide, and publishes it in
manual_location- Parameters
docs_folder (str) – The path to sphinx’s docs folder, where the tests will be run; by default
./docs/work_dir (str) – The current Working Directory; by default
prechecks_done (bool) – Whether the prechecks present in ~CanGraph.deploy.make_sphinx_prechecks have already been made
custom_domain (str) – A custom domain to deploy de docs to.
manual_location (str) – The location (including filename) of the finalised PDF manual, relative to the location of the script
- Returns
Whether the prechecks have already been done; always True if the function is run
- Return type
- deploy_webdocs(docs_folder='./docs/', work_dir='.', prechecks_done=False, custom_domain=None)[source]#
Generates the HTML web docs, and publishes it both to Github and Codeberg pages
- Parameters
docs_folder (str) – The path to sphinx’s docs folder, where the tests will be run; by default
./docs/work_dir (str) – The current Working Directory; by default
prechecks_done (bool) – Whether the prechecks present in ~CanGraph.deploy.make_sphinx_prechecks have already been made
custom_domain (str) – A custom domain to deploy de docs to.
- Returns
Whether the prechecks have already been done; always True if the function is run
- Return type
Note
For
custom_domainto work, please configure your DNS records apparentlyNote
modules.rstis not removed, but it is correctly ignored in conf.py
- git_push(path_to_repo, remote_names, commit_message, force=False)[source]#
Pushes the current repo’s state and current branch to a remote git repository
- Parameters
path_to_repo (str) – The path to the local
.gitfolderremote_names (list or str) – The names of the remote to which we want to commit, which must be previously configured (see
CanGraph.setup.setup_git). e.g.: [“github”, “codeberg”]commit_message (str) – The Git Commit Message for the current repo’s state
force (bool) – Whether to force the commit (necessary if you are resetting the HEAD)
Note
gitpythonis not good at managing complex commit messages (i.e. those with a Subject and a Body). If you want to add one of those, please, use\nas the separator; the function will take care of the restSee also
The approach taken hare was inspired by StackOverflow #41836988
CanGraph.main module#
A python module that leverages the functions present in the miscelaneous
module and all other subpackages to annotate metabolites using a graph format and Neo4J,
and then provides an GraphML export file.
CanGraph.main Usage#
To use this module:
A python utility to study and analyse cancer-associated metabolites using knowledge graphs
usage: python3 main.py [-h] [-c] [-n] [-s] [-w] [-i] --query QUERY
[--dbfolder DBFOLDER] [--results RESULTS]
[--adress ADRESS] [--username USERNAME]
[--password PASSWORD]
Named Arguments#
- -c, --check_args
Checks if the rest of the arguments are OK, then exits
- -n, --noindex
Runs the program checking each file one-by-one, instead of using a JSON index
- -s, --similarity
Deactivates the import of information based on Structural Similarity.This might dramatically increase processing time; default is True.
- -w, --webdbs
Activates import of information based on web databases.This might dramatically increase processing time; default is True.
- -i, --interactive
tells the script if it wants interaction from the user and more information shown to them; similar to –verbose
- --query
The location of the CSV file in which the program will search for metabolites
- --dbfolder
The folder indicated to
`setup.py`as the one where your databases will be stored; default is./DataBases- --results
The folder where the resulting GraphML exports will be stored; default is
./Results- --adress
the URL of the database, in neo4j:// or bolt:// format
- --username
the username of the neo4j database in use
- --password
the password for the neo4j database in use. NOTE: Since passed through bash, you may need to escape some chars
You may find more info in the package’s README.
Note
For this program to work, the Git environment has to be set up first.
You can ensure this by using: CanGraph.setup.setup_git
CanGraph.main Functions#
This module is comprised of:
- add_mesh_and_metanetx(driver)[source]#
Add MeSH Term IDs, Synonym relations and Protein interactions to existing nodes using MeSH and MetaNetX Also, adds Kegg Pathway IDs
- Parameters
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
- annotate_using_wikidata(driver)[source]#
Once we finish the search, we annotate the nodes added to the database using WikiData
- Parameters
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
Todo
When fixing queries, fix the main subscript also
- args_parser()[source]#
Parses the command line arguments into a more usable form, providing help and more
- Returns
A dictionary of the different possible options for the program as keys, specifying their set value. If no command-line arguments are provided, the help message is shown and the program exits.
- Return type
Note
Note that, in Google Docstrings, if you want a multi-line
Returnscomment, you have to start it in a different line :(Note
The return must be of type
argparse.ArgumentParserfor theargparsedirective to work and auto-gen docsNote
By using
argparse.constinstead ofargparse.default, the check_file function will check “” (the current dir, always exists) if the arg is not provided, not breaking the function; if it is, it checks it.
- build_from_file(filepath, Neo4JImportPath, driver)[source]#
Imports a given metabolite from a sigle-metabolite containing file by checking its type and calling the appropriate import functions.
- Parameters
filepath (str) – The path to the file in which will be imported
Neo4JImportPath (str) – The path which Neo4J will use to import data
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
- Returns
This function does not provide a particular return, but rather imports the requested file
Note
The
filepathmay be absolute or relative, but it is transformed to a relativerelpathin order to remove possible influence of higher-name folders in the import type selection. This is also why the condition is stated as a big “if/elif/else” instead of a series of “ifs”
- find_reasons_to_import_all_files(filepath, similarity, chebi_ids, names, hmdb_ids, inchis, mesh_ids)[source]#
Finds reasons to import a metabolite given a candidate filepath with one metabolite per file and a series of lists containing all synonyms of the values considered reasons for import
- Parameters
filepath (str) – The path to the file in which we will search for reasons to import
similarity (bool) – Whether to use similarity as a measure to import or not
chebi_ids (list) – A list of all the ChEBI_ID which are considered a reason to import
names (list) – A list of all the Name which are considered a reason to import
hmdb_ids (list) – A list of all the HMDB_ID which are considered a reason to import
inchis (list) – A list of all the InChI which are considered a reason to import
mesh_ids (list) – A list of all the MeSH_ID which are considered a reason to import
- Returns
A list of the methods that turned out to be valid for import, such as Name, ChEBI_ID…
- Return type
- find_reasons_to_import_inchi(query, subject)[source]#
Takes two chains of text and finds if the
queryis present in thesubject, or if there are molecules common between them with at least 95% similarity- Parameters
- Returns
A dict with each query as a key and the reason to import it as value, if there is one.
- Return type
See also
This approach was taken from Chemistry StackExchange #82144
Note
Since this is a one-to-one comparison, subject and query can be used interchangeably; however, bear in mind that only the query can be provided as a list
- import_based_on_all_files(all_files, Neo4JImportPath, driver, similarity, chebi_ids, names, hmdb_ids, inchis, mesh_ids)[source]#
A function that searches inside a series of lists, provided as arguments, and imports the metabolites matching those present in them iterating over a list of files which may contain relevant information to be imported
- Parameters
all_files (list) – A list of all the posible files where we want to look for info
Neo4JImportPath (str) – The path which Neo4J will use to import data
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
similarity (bool) – Whether to use similarity as a measure to import or not
chebi_ids (list) – A list of all the ChEBI_ID which are considered a reason to import
names (list) – A list of all the Name which are considered a reason to import
hmdb_ids (list) – A list of all the HMDB_ID which are considered a reason to import
inchis (list) – A list of all the InChI which are considered a reason to import
mesh_ids (list) – A list of all the MeSH_ID which are considered a reason to import
- import_based_on_index(databasefolder, Neo4JImportPath, driver, similarity, chebi_ids, names, hmdb_ids, inchis, mesh_ids)[source]#
A function that searches inside a series of lists, provided as arguments, and imports the metabolites matching those present in them using a JSON file to map the bits of the databases where the relevant information lies
- Parameters
databasefolder (str) – The main folder where all the databases we will be using are to be found There must be an index.json file located in
databasefolder/index.jsonNeo4JImportPath (str) – The path which Neo4J will use to import data
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
similarity (bool) – Whether to use similarity as a measure to import or not
chebi_ids (list) – A list of all the ChEBI_ID which are considered a reason to import
names (list) – A list of all the Name which are considered a reason to import
hmdb_ids (list) – A list of all the HMDB_ID which are considered a reason to import
inchis (list) – A list of all the InChI which are considered a reason to import
mesh_ids (list) – A list of all the MeSH_ID which are considered a reason to import
- improve_search_terms(driver, chebi_ids, names, hmdb_ids, inchis, mesh_ids)[source]#
Improves the search terms already provided to the CanGraph programme by processing the text stings and finding synonyms in various platforms
- Parameters
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
chebi_ids (str) – A string of “;” separated values of all the ChEBI_ID representing the current metabolite
names (list) – A string of “;” separated values of all the Name representing the current metabolite
hmdb_ids (list) – A string of “;” separated values of all the HMDB_ID representing the current metabolite
inchis (list) – A string of “;” separated values of all the InChI representing the current metabolite
mesh_ids (list) – A string of “;” separated values of all the MeSH_ID representing the current metabolite
- Returns
A list containing [ chebi_ids, names, hmdb_ids, inchis, mesh_ids ], with all their synonyms
- Return type
- improve_search_terms_with_cts(query, query_type, chebi_ids, names, hmdb_ids, inchis, mesh_ids)[source]#
Improves the search terms already provided to the CanGraph programme by using The Chemical Translation Service to find synonyms in IDs
- Parameters
query (str) – The term we are currently querying for
query_type (str) – The kind of query to search; one of [“ChEBI_ID”, “HMDB_ID”, “Name”, “InChI”, “MeSH_ID”]
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
chebi_ids (str) – A string of “;” separated values of all the ChEBI_ID representing the current metabolite
names (list) – A string of “;” separated values of all the Name representing the current metabolite
hmdb_ids (list) – A string of “;” separated values of all the HMDB_ID representing the current metabolite
inchis (list) – A string of “;” separated values of all the InChI representing the current metabolite
mesh_ids (list) – A string of “;” separated values of all the MeSH_ID representing the current metabolite
- Returns
A list containing [ chebi_ids, names, hmdb_ids, inchis, mesh_ids ], with all their synonyms
- Return type
- improve_search_terms_with_metanetx(query, query_type, driver, chebi_ids, names, hmdb_ids, inchis, mesh_ids)[source]#
Improves the search terms already provided to the CanGraph programme by using the MetaNetX web service to find synonyms in IDs
- Parameters
query (str) – The term we are currently querying for
query_type (str) – The kind of query to search; one of [“ChEBI_ID”, “HMDB_ID”, “Name”, “InChI”, “MeSH_ID”]
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
chebi_ids (str) – A string of “;” separated values of all the ChEBI_ID representing the current metabolite
names (list) – A string of “;” separated values of all the Name representing the current metabolite
hmdb_ids (list) – A string of “;” separated values of all the HMDB_ID representing the current metabolite
inchis (list) – A string of “;” separated values of all the InChI representing the current metabolite
mesh_ids (list) – A string of “;” separated values of all the MeSH_ID representing the current metabolite
- Returns
A list containing [ chebi_ids, names, hmdb_ids, inchis, mesh_ids ], with all their synonyms
- Return type
- link_to_original_data(item_type, item, import_based_on)[source]#
Links a recently-imported metabolite to the original data (that which caused it to be imported) by creating an
ÒriginalMetabolitenode that is(n)-[r:ORIGINALLY_IDENTIFIED_AS]->(a)related to the imported data- Parameters
tx (neo4j.Session) – The session under which the driver is running
item_type (str) – The property to match in the Neo4J DataBase
item (dict) – The value of property
`item_type`import_based_on (list) – A list of the methods that turned out to be valid for import, such as Name, ChEBI_ID…
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- main()[source]#
The function that executes the code
Note
This function disables rdkit’s log messages, since rdkit seems to dislike the way some of the InChI strings it is getting from the databases are formatted
Todo
CAMBIAR NOMBRE A LOS MESH PARA INDICAR EL TIPO. AÑADIR NAME A LOS WIKIDATA
Todo
FIX THE REPEAT TRANSACTION FUNCTION
Todo
Match partial InChI based on DICE-MACCS
Todo
QUE FUNCIONE -> ACTUALMENTE ESTA SECCION RALENTIZA MAZO
Todo
CHECK APOC IS INSTALLED
Todo
FIX MAIN
Todo
MERGE BY INCHI, METANETX ID
Todo
Fix find_protein_interactions_in_metanetx
Todo
Mover esa funcion de setup a misc
Todo
EDIT conf.py
Todo
Document the following Schema Changes: * For Subject, we have a composite PK: Exposome_Explorer_ID, Age, Gender e Information * Now, more diseases will have a WikiData_ID and a related MeSH. This will help with networking. And, this diseases dont even need to be a part of a cancer! * The Gene nodes no longer exist in the full db? -> They do
CanGraph.miscelaneous module#
A python module that provides a collection of functions to be used across the different scripts present in the CanGraph package, with various, useful functionalities
- call_db_schema_visualization()[source]#
Shows the DB Schema. This function is intended to be run only in Neo4J’s console, since it produces no output when called from the driver.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
Todo
Make it download the image
- check_file(filepath)[source]#
Checks for the presence of a file or folder. If it exists, it returns the filepath; if it doesn’t, it raises an
argparse.ArgumentTypeError, which tells argparse how to process file exclussionNote
Perhaps its not ideal, but I will be using this also to check for file existence throughout the CanGraph project, although the error type might not be correct
- check_neo4j_protocol(string)[source]#
Checks that a given
stringstarts with any of the protocols accepted by theneo4j.Driver- Parameters
string (str) – A string, which will normally represent the neo4j adress
- Returns
The same string that was provided as an argument (required by
argparse.ArgumentParser)- Return type
- Raises
argparse.ArgumentTypeError – If the string is not of the correct protocol
- clean_database()[source]#
A CYPHER query that gets all the nodes in a Neo4J database and removes them, in transactions of 100 rows to alleviate memory load
- Returns
A text chain that represents the CYPHER query with the desired output. This can be run using:
neo4j.Session.run- Return type
Note
This is an autocommit transaction. This means that, in order to not keep data in memory (and make running it with a huge amount of data) more efficient, you will need to add
`:auto `when calling it from the Neo4J browser, or call it as usingneo4j.Session.runfrom the driver.
- connect_to_neo4j(port='bolt://localhost:7687', username='neo4j', password='neo4j')[source]#
A function that establishes a connection to the neo4j server and returns a
Driverinto which transactions can be passed- Parameters
- Returns
An instance of Neo4J’s Bolt Driver that can be used
- Return type
Note
Since this is a really short function, this doesn’t really simplify the code that much, but it makes it much more re-usable and understandable
- countlines(start, header=True, lines=0, begin_start=None)[source]#
A function that counts all the lines of code present in a given directory; useful to show off in Sphinx Docs
- Parameters
- Returns
The number of lines present in
start- Return type
See also
This function was taken from StackOverflow #38543709
- create_n10s_graphconfig()[source]#
A CYPHER query that creates a neosemantics (n10s) constraint to hold all the RDF we will import.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
See also
More information on this approach can be found in Neosemantics’ 101 Guide and in Neo4J’s guide on how to import data from Wikidata , where this approach was taken from
Deprecated since version 0.9: Since we are importing based on apoc.load.jsonParams, this is not needed anymore
- download_and_unzip(url, folder)[source]#
Downloads and unzips a given Zipfile from the internet; useful for databases which provide zip access.
- Parameters
- Returns
This function downloads and unzips the file in the desired folder, but does not produce any particular return.
See also
Code snippets for this function were taken from Shyamal Vaderia’s Github and from StackOverflow #32123394
- export_graphml(exportname)[source]#
Exports a Neo4J graph to GraphML format. The graph will be exported to Neo4JImportPath
- Parameters
exportname (str) – The name for the exported file, which will be saved under ./Neo4JImportPath/
- Returns
- A Neo4J connexion to the database that exports the file, using batch optimizations and
smaller batch sizes to try to keep the impact on memory use low
- Return type
Note
for this to work, you HAVE TO have APOC availaible on your Neo4J installation
- get_import_path(driver)[source]#
A function that runs an autocommit transaction to get Neo4J’s Import Path
Note
By doing the Neo4JImportPath search this way (in two functions), we are able to run the query as a :obj: execute_read, which, unlike autocommit transactions, allows the query to be better controlled, and repeated in case it fails.
- Parameters
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
- Returns
Neo4J’s Import Path, i.e., where Neo4J will pick up files to be imported using the
`file:///`schema- Return type
- import_graphml(importname)[source]#
Imports a GraphML file into a Neo4J graph. The file has to be located in Neo4JImportPath
- Parameters
importname (str) – The name for the file to be imported, which must be under ./Neo4JImportPath/
- Returns
- A Neo4J connexion to the database that imports the file, using batch optimizations and
smaller batch sizes to try to keep the impact on memory use low
- Return type
Note
for this to work, you HAVE TO have APOC availaible on your Neo4J installation
- kill_neo4j(neo4j_home='neo4j')[source]#
A simple function that kills any process that was started using a cmd argument including “neo4j”
- Parameters
neo4j_home (str) – the installation directory for the
neo4jprogram; by default,neo4j
Warning
This function may unintendedly kill any command run from the
neo4jfolder. This is unfortunate, but the creation of this function was essential given thatneo4j stopdoes not work properly; instead of dying, the process lingers on, interfering withfind_neo4j_installation_statusand hindering the main program
- manage_transaction(tx, driver, num_retries=10, neo4j_home='neo4j', **kwargs)[source]#
A function that repeats transactions whenever an error is found. This may make an incorrect script unnecessarily repeat; however, since the error is printed, one can discriminate those out, and the function remains helpful to prevent SPARQL Read Time-Outs.
It will also re-start neo4j in case it randomly dies while executing a query.
- Parameters
tx (str) – The transaction that we desire to run, specified as a CYPHER query
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
num_retries (int) – The number of times that we wish the transaction to be retried
neo4j_home (str) – the installation directory for the
neo4jprogram; by default,neo4j**kwargs – Any number of arbitrary keyword arguments
- Raises
Exception – An exception telling the user that the maximum number of retries has been exceded, if such a thing happens
- Returns
The response from the Neo4J Database
- Return type
Note
This function does not accept args, but only kwargs (named keyword arguments). Thus, if you wish to add a parameter (say,
number, you should add it as:number=33
- merge_duplicate_nodes(node_types, node_property, optional_condition='', more_props='')[source]#
Removes any two nodes of any given
`node_type`with the same`condition`.- Parameters
node_types (str) – The labels of the nodes that will be selected for merging; i.e.
n:Fruit OR n:Vegetablenode_property (str) – The node properties used for collecting, if not using all properties.
optional_condition (str) – An optional Neo4J Statement, starting with “AND”, to be added after the
WHEREclause.
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Warning
When using, take good care on how the keys names are written: sometimes, if a key is not present, all nodes will be merged!
- old_sleep_with_counter(seconds, step=20, message='Waiting...')[source]#
A function that waits while showing a cute animation, but without using the ``alive_progress` module
Note
This function interacts weirdly with slurn; I’d recommend to not use it on the HPC
- purge_database(driver, method=['merge', 'delete'])[source]#
A series of commands that purge a database, removing unnecessary, duplicated or empty nodes and merging those without required properties. This has been converted into a common function to standarize the ways the nodes are merged.
- Args:
driver (neo4j.Driver): Neo4J’s Bolt Driver currently in use method (list): The part of the function that we want to execute; if [“delete”], only call
queries that delete nodes; if [“merge”], only call those that merge; if both, do both
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
Warning
When modifying, take good care on how the keys names are written: with
merge_duplicate_nodes, sometimes, if a key is not present, all nodes will be merged!
- remove_ExternalEquivalent()[source]#
Removes all nodes of type: ExternalEquivalent from he DataBase; since this do not add new info, one might consider them not useful.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- remove_duplicate_relationships()[source]#
Removes duplicated relationships between ANY existing pair of nodes.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
Only deletes DIRECTED relationships between THE SAME nodes, combining their properties
See also
This way of working has been taken from StackOverflow #18724939
- remove_n10s_graphconfig()[source]#
Removes the “_GraphConfig” node, which is necessary for querying SPARQL endpoints but not at all useful in our final export
- Parameters
tx (neo4j.Session) – The session under which the driver is running
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Deprecated since version 0.9: Since we are importing based on apoc.load.jsonParams, this is not needed anymore
- restart_neo4j(neo4j_home='neo4j')[source]#
A simple function that (re)starts a neo4j server and returns its bolt adress
- Parameters
neo4j_home (str) – the installation directory for the
neo4jprogram; by default,neo4j
Note
Re-starting is better than starting, as it tries to kills old sessions (a task at which it fails miserably, thus the need for
kill_neo4j), and, most importantly, because it returns the currently used bolt port
- sleep_with_counter(seconds, step=20, message='Waiting...')[source]#
A function that waits while showing a cute animation
- split_csv(filename, folder, sep=',', sep_out=',', startFrom=0, withStepsOf=1)[source]#
Splits a given .csv/tsv file in n smaller csv files, one for each row on the original file, so that it does not crash when processing it. It also allows to start reading from
`startFrom`lines- Parameters
- Returns
The number of files that have been produced from the original
- Return type
Warning
The original file will be removed
- split_xml(filepath, splittag, bigtag)[source]#
Splits a given .xml file in n smaller XML files, one for each
splittagsection that is pressent in the original file, which should be of typebigtag. For example, we might have an<hmdb>file which we want to slit based on the<metabolite>items therein contained. Ths is so that Neo4J does not crash when processing it.- Parameters
- Returns
The number of files that have been produced from the original
- Return type
Warning
The original file will be removed
- untargz(file_path, folder)[source]#
Untargzs a file present at a given
file_pathinto a givenfolder
CanGraph.setup module#
A python module that prepares the local environment, to be able to run the main
and deploy functions. This can be either run in an interactive way, requiring
user input; or in a automatic way, in order to pre-configure things, for example, if you are using
the singularity package
CanGraph.setup Usage#
To use this module:
A python module that prepares the local environment, to be able to run the CanGraph.main and CanGraph.deploy functions.
usage: python3 setup.py [-h] [-i] [-a] [--dbfolder [DBFOLDER]] [--git [GIT]]
[--requirements [REQUIREMENTS]] [-n [NEO4J]]
[--neo4j_username [NEO4J_USERNAME]]
[--neo4j_password [NEO4J_PASSWORD]]
Named Arguments#
- -i, --interactive
tells the script if it wants interaction from the user and information shown to them; similar to –verbose
- -a, --all
runs all the options below at once; equivalent to -dgnr; it DOES NOT activate the interactive mode
- --dbfolder
set up the databases from which the program will pull its info using the provided folder
- --git
prepare the git environment for the deploy script using the provided git folder
- --requirements
installs all the requirements needed for all the possible options from the given requirements file
- -n, --neo4j
set up the neo4j local environment, to run from the provided folder
- --neo4j_username
the username for the neo4j database
- --neo4j_password
the password for the neo4j database
CanGraph.setup Functions#
This module is comprised of:
- args_parser()[source]#
Parses the command line arguments into a more usable form, providing help and more
- Returns
A dictionary of the different possible options for the program as keys, specifying their set value. If no command-line arguments are provided, the help message is shown and the program exits.
- Return type
Note
Note that, in Google Docstrings, if you want a multi-line
Returnscomment, you have to start it in a different line :(Note
The return must be of type
argparse.ArgumentParserfor theargparsedirective to work and auto-gen docsNote
The
--all```option has to be adressed outside of this function in order to not mess up the ``argparsedirective in sphinxNote
By using
argparse.constinstead ofargparse.default, the check_file function will check “” (the current dir, always exists) if the arg is not provided, not breaking the function; if it is, it checks it.
- change_neo4j_password(new_password, old_password='neo4j', user='neo4j', database='system', neo4j_home='neo4j')[source]#
Changes the neo4j password for user
user, fromold_passwordtonew_password, by using a simple query in cypher-shell- Parameters
neo4j_home (str) – the installation directory for the
neo4jprogram; by default,neo4jnew_password (str) – the new password for the database
old_password (str) – the old password for the database, needed for identification.
user (str) – the user for which the password is being changed.
database (str) – the name of the database for which we want to modify the password. By default, it is
system, since Neo4J’s community edition only allows for one database
Warning
DO NOT REMOVE THE TRY-EXCEPT BLOCK THAT ATTEMPTS TO CONNECT TO NEO4J: It somehow magically maked the passsword change work. IT WILL NOT WORK IF THAT LINES ARE NOT PRESENT
- check_exposome_files(databasefolder='./DataBases')[source]#
Checks for the presence of all the files that should be in “
databasefolder/ExposomeExplorer”` for the ExposomeExplorer part of the script to run
- configure_neo4j(neo4j_home='neo4j')[source]#
Modifies the Neo4J conf file according to some recommendations provided by
memrec, neo4j’s memory recommendator. It also enables the Awesome Procedures On Cypher (APOC) plugin from Neo4j Labs, and enables other basic confs such as file export and import or bigger timeouts- Parameters
neo4j_home (str) – the installation directory for the
neo4jprogram; by default,neo4j
Note
In order to make the setup more consistent, this function also forces the Neo4JImportPath (
dbms.directories.import) to be presented in an absolute way, instead of being relative toneo4j_home
- final_message(interactive=False)[source]#
Prompts the user with a final message.
- Parameters
interactive (bool) – Whether the session is set to be interactive or not
- find_neo4j_installation_status(neo4j_home='neo4j', neo4j_username='neo4j', neo4j_password='neo4j')[source]#
Finds the installation status of Neo4J by trying to use it normally, and analyzing any thrown exceptions
- Parameters
- Returns
A list of two booleans: whether neo4j exists at
neo4j_home, and whether the supplied credentials are valid or not- Return type
- initial_message()[source]#
Prompts the user with an initial message if the session is set to be interactive.
- Parameters
interactive (bool) – Whether the session is set to be interactive or not
- install_neo4j(neo4j_home='neo4j', interactive=False, version='4.4.0')[source]#
Installs the neo4j database program in the
neo4j_homefolder, by getting it from the internet according to the Operating System the script is been run in (aims for multi-platform!)
- install_packages(requirements_file=None, package_name=None, interactive=False)[source]#
Automates installing packages using PIP
- Parameters
- Raises
ValueError – If neither a
requirements_filenor apackage_nameis provided
- setup_database_index(databasefolder='./DataBases')[source]#
Prepares the index file for all the databases present in the
databasefolderfolder, which will helpfully reduce processing time a lot
- setup_databases(databasefolder='./DataBases', interactive=False)[source]#
Set Up the
databasefolderfrom where themainscript will take its data. It does so by creating or removing and re-creating thedatabasefolder, and putting inside it, or asking/checking if the user has put inside, the necessary files
- setup_drugbank(databasefolder='./DataBases', interactive=False)[source]#
Sets up the files relative to the SMPDB database in the
databasefolder, splitting them for easier processing later on.- Parameters
- Returns
True if everything went okay; False otherwise. If False, DrugBank should not be used as a data source
- Return type
Warning
When updating the DrugBank DataBase Version, please edit this function to reflect the correct number of files
- setup_exposome(databasefolder='./DataBases', interactive=False)[source]#
Sets up the files relative to the Exposome Explorer database in the
databasefolder, splitting them for easier processing later on. If the session is set to be interactive, the user will be given time to add the files themselves; if not, the full suite of necessary files will be checked for their presence indatabasefolderThen, the “components” file will be splitted into one record oer line, as
mainrequires- Parameters
- Returns
True if everything went okay; False otherwise. If False, Exposome-Explorer should not be used as a data source
- Return type
Warning
When updating the Exposome Explorer DataBase Version, please edit
check_exposome_filesto reflect the correct number of files
- setup_folders(databasefolder='./DataBases', interactive=False)[source]#
Creates the
databasefolderif it does not exist. If it does, it either asks before overwriting ininteractivemode, or directly overwrites in auto mode.- Parameters
- Raises
ValueError – If the Databases folder already exists (so as not to overwrite)
- Returns
True if successful, False otherwise.
- Return type
- setup_git(path_to_repo='.git')[source]#
Set Up the Git environment for the
deployscript. It does so by removing any existing remotes and setting two new ones: github and codeberg, with their respective branches- Parameters
path_to_repo (str) – The path to the Git repo; by default,
.git
- setup_hmdb(databasefolder='./DataBases')[source]#
Sets up the files relative to the HMDB database in the
databasefolder, splitting them for easier processing later on.- Parameters
databasefolder (str) – The main folder where all the databases we will be using are to be found
- Returns
True if everything went okay; False otherwise. If False, DrugBank should not be used as a data source
- Return type
Warning
When updating the Exposome Explorer DataBase Version, please edit
check_exposome_filesto reflect the correct number of files
- setup_neo4j(neo4j_home='neo4j', neo4j_username='neo4j', neo4j_password='neo4j', interactive=False)[source]#
Sets ups the neo4j environment in
neo4j_home, so that the functions inmaincan propperly function. Using the functions present in this module, it finds if neo4j is installed with default credentials, and, if not, it installs it, changing the default password to a new one, and returning its value- Parameters
neo4j_home (str) – the installation directory for the
neo4jprogram; by default,neo4jneo4j_username (str) – the username for the neo4j database; by default
neo4jneo4j_password (str) – the password for the neo4j database; by default
neo4jinteractive (str) – tells the script if it wants interaction from the user and information shown to them
- Returns
The password that was set up for the new neo4j database. This is also written to .neo4jpassword
- Return type
Note
This has been designed to be used with a
neo4j_homelocated in the WorkDir, but can be used in any other location with read/write access, or even withapt installinstalled versions! Just find itsneo4j_home, make sure it has r/w access, and provide it to the program!Note
If no neo4j_password is provided or if neo4j_password = “neo4j”, the function will check for a previously created “.neo4jpassword” file, signalling a possible pre-existing database with known credentials
- setup_smpdb(databasefolder='./DataBases')[source]#
Sets up the files relative to the SMPDB database in the
databasefolder, splitting them for easier processing later on.
- update_neo4j_confs(key, value, conf_file='neo4j/conf/neo4j.conf')[source]#
Updates a preference on neo4j’s
conf_file, given its name (key) and its expectedvalueIf a preference is set with a value other thankey, said value will be overwritten; if it is commented, it will be uncommented (thanks to regex!)
