CanGraph.GraphifySMPDB package
In this page
CanGraph.GraphifySMPDB package#
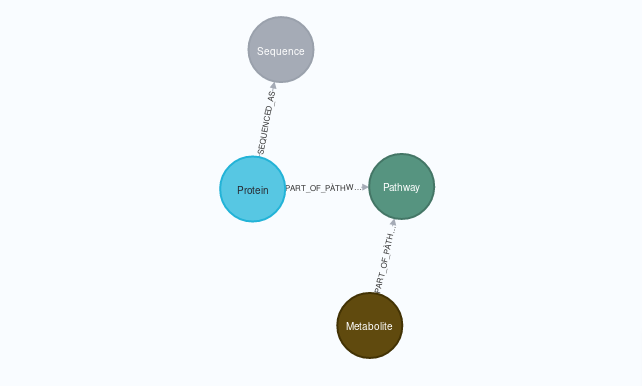
This script, created as part of my Master’s Intenship at IARC, transitions the Small Molecule Pathway Database (a high quality database containing associations betweem metabolites, proteins and metabolomic pathways) to Neo4J format in an automated way, providing an export in GraphML format.
To run, it uses alive_progress to generate an interactive progress bar (that shows the script is still running through its most time-consuming parts) and the neo4j python driver. This requirements can be installed using: pip install -r requirements.txt.
To run the script itself, use:
python3 main.py neo4jadress databasename databasepassword
where:
neo4jadress: is the URL of the database, in neo4j:// or bolt:// format
databasename: the name of the database in use. If using the free version, there will only be one database per project (neo4j being the default name); if using the pro version, you can specify an alternate name here
databasepassword: the passowrd for the databasename DataBase. Since the arguments are passed by BaSH onto python3, you might need to escape special characters
NOTE: The files will be downloaded to ./csvfolder, so please run the script somewhere you have read/write permissions
An archived version of this repository that takes into account the gitignored files can be created using: git archive HEAD -o ${PWD##*/}.zip
Important Notices on SMPDB#
Please ensure you have internet access, enough espace in your hard drive (around 5 GB) and read-write access in
./csvfolder. The files needed to build the database will be stored there.Since the “Structure” files at smpdb.ca seemed to be super complicated to import, we decided against doing so. However, regarding the future, they shouldn’t be overlooked, as they might include useful info
The “PW ID” column from the “Pathways” table, and the “Pathway Subject” from the “Metabolite” tables may correlate (both start with PW). However, they seem to use different formats, so we have decided against including them in the Final Database
The package consists of the following modules:
CanGraph.GraphifySMPDB.build_database module#
A python module that provides the necessary functions to transition the SMPDB database to graph format,
either from scratch importing all the nodes (as showcased in CanGraph.GraphifySMPDB.main) or in a case-by-case basis,
to annotate existing metabolites (as showcased in CanGraph.main).
- add_metabolites(filename)[source]#
Adds “Metabolite” nodes to the database, according to individual CSVs present in the SMPDB website
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the CSV file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
Some of the node’s properties might be set to “null” (important in order to work with it)
Note
This database clearly differentiates Metabolites and Proteins, so no overlap is accounted for
- add_pathways(filename)[source]#
Adds “Pathways” nodes to the database, according to individual CSVs present in the SMPDB website Since this is done after the creation of said pathways in the last step, this will most likely just annotate them.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the CSV file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Todo
This file is really big. It could be divided into smaller ones.
- add_proteins(filename)[source]#
Adds “Protein” nodes to the database, according to individual CSVs present in the SMPDB website
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the CSV file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
Some of the node’s properties might be set to “null” (important in order to work with it)
Note
This database clearly differentiates Metabolites and Proteins, so no overlap is accounted for
Todo
Why is the SMPDB_ID property called like that and not SMDB_ID?
Warning
Since no unique identifier was found, CREATE had to be used (instead of merge). This might create duplicates. which should be accounted for.
- add_sequence(seq_id, seq_name, seq_type, seq, seq_format='FASTA')[source]#
Adds “Pathways” nodes to the database, according to the sequences presented in FASTA files from the SMPDB website
- Parameters
tx (neo4j.Session) – The session under which the driver is running
seq_id (str) – The UniProt Database Identifier for the sequence that is been imported
seq_name (str) – The Name (i.e. FASTA header) of the Sequence that is been imported
seq_type (bool) – The type of the sequence; can be either of [“DNA”, “PROT”]
seq (str) – The seuqnce that is been imported; a text chain of nucleotides or aminoacids, identified by their acronyms
seq_format (str) – The format the sequence is provided under; default is “FASTA”, but its optional
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- build_from_file(filepath, Neo4JImportPath, driver, filetype)[source]#
A function able to build a portion of the SMPDB in graph format, provided that one CSV is supplied to it. This CSVs are downloaded from the website, and can be presented either as the full file, or as a splitted version of it, with just one item per file (which is recommended due to memory limitations)
Since file title represents a different pathway, the function automatically picks up and import the relative pathway node.
- Parameters
filepath (str) – The path to the current file being imported
Neo4JImportPath (str) – The path from which Neo4J is importing data
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
filetype (bool) – The type of file being imported; one of ether [“Metabolite”, “Protein”]- If the file is a FASTA sequence store, this will be auto-detected.
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
Note
Since this adds a ton of low-resolution nodes, maybe have this db run first?
CanGraph.GraphifySMPDB.main module#
A python module that leverages the functions present in the build_database
module to recreate the SMPDB database using a graph format and Neo4J,
and then provides an GraphML export file.
Please note that, to work, the functions here pre-suppose you have internet access, which will be used to download
HMDB’s CSVs under `./csvfolder/` (please ensure you have read-write access there).
For more details on how to run this script, please consult the package’s README
- import_genomic_seqs()[source]#
Imports the
`smpdb_gene.fasta`file from the SMPDB Database. The function assumes the file is availaible at`./csvfolder/smpdb_gene.fasta`- Parameters
Neo4JImportPath (str) – The path from which Neo4J is importing data
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
- import_metabolites(filename, Neo4JImportPath)[source]#
Imports “Metabolite” files from the SMPDB Database. The function assumes
`filename`is availaible at`./csvfolder/smpdb_metabolites/`
- import_pathways(Neo4JImportPath)[source]#
Imports the
`smpdb_pathways.csv`file from the SMPDB Database. The function assumes the file is availaible at`./csvfolder/smpdb_gene.fasta`- Parameters
Neo4JImportPath (str) – The path from which Neo4J is importing data
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
- import_proteic_seqs()[source]#
Imports the
`smpdb_protein.fasta`file from the SMPDB Database. The function assumes the file is availaible at`./csvfolder/smpdb_protein.fasta`- Parameters
Neo4JImportPath (str) – The path from which Neo4J is importing data
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
