CanGraph.QueryWikidata package
In this page
CanGraph.QueryWikidata package#
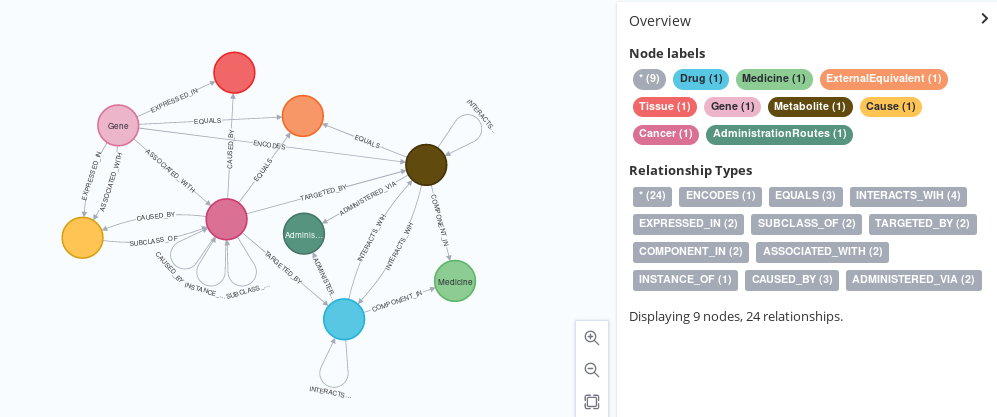
This script, created as part of my Master’s Intenship at IARC, imports nodes from the WikiData SPARQL Service, creating a high-quality representation of the data therein. Although wikidata is manually curated using the Wiki principles, some publications have found it might be a good source of information for life sciences, specially due to the breadth of information it contains. It also provides an export in GraphML format.
To run, it uses alive_progress to generate an interactive progress bar (that shows the script is still running through its most time-consuming parts) and the neo4j python driver. This requirements can be installed using: pip install -r requirements.txt.
To run the script itself, use:
python3 build_database.py neo4jadress username databasepassword
where:
neo4jadress: is the URL of the database, in neo4j:// or bolt:// format
username: the username for your neo4j instance. Remember, the default is neo4j
password: the passowrd for your database. Since the arguments are passed by BaSH onto python3, you might need to escape special characters
Please note that there are two kinds of functions in the associated code: those that use python f-strings, which themselves contain text that cannot be directly copied into Neo4J (for instance, double brackets have to be turned into simple brackets) and normal multi-line strings, which can. This is because f-strings allow for variable customization, while normal strings dont.
An archived version of this repository that takes into account the gitignored files can be created using: git archive HEAD -o ${PWD##*/}.zip
Finally, please node that the general philosophy and approach of the queries have been taken from Towards Data Science, a genuinely useful web site.
Important Notices on WikiData#
Please ensure you have internet access, which will be used to connect to Wikidata’s SPAQL endpoint and gather the necessary info.
As Neo4J can run out of “Java Heap Space” if the number of nodes/properties to add is too high, the script has been divided in order to minimize said number: for instance, only nodes with a
wikidata_idending in a given number from 0 to 9 are processed at a time. This does not decrease performance, since these nodes would have been processed nontheless, but makes the script more reliable.What does impact performance, however, is having different functions for adding cancers, drugs, metabolites, etc, instead of having just one match for each created cancer node. This makes WikiData have to process more queries that are less heavy, which makes it less likely to time-out, but causes the script to run more slowly.
The Neo4J server presents a somewhat unstable connection that is sometimes difficult to keep alive, as it tends to be killed by the system when you so much as look at it wrong. To prevent this from happening, you are encouraged to assign a high-priority to the server’s process by using the
niceorrenicecommands in Linux (note that the process will be called “Java”, not “Neo4J”)Another measure taken to prevent Neo4J’s unreliability from stopping the script is the
misc.manage_transactionfunction, which insists a given number of times until either the problem is fixed or the error persists. This is because Neo4J tends to: random disconnects, run out of java heap space, explode… and WikiData tends to give server errors, have downtimes during the 14+ hours the script takes to run, etc.The data present in the “graph.graphml” file comes from WikiData, and was provided by this service free of charge and of royalties under the permissive CC-0 license.
The package consists of the following modules:
CanGraph.QueryWikidata.build_database module#
A python module that provides the necessary functions to transition selected parts of the Wikidata database to graph format,
either from scratch importing all the nodes (as showcased in CanGraph.QueryWikidata.main) or in a case-by-case basis,
to annotate existing metabolites (as showcased in CanGraph.main).
Note
You may notice some functions here present the **kwargs arguments option.
This is in order to make the functions compatible with the
CanGraph.miscelaneous.manage_transaction function, which might send back a variable
number of arguments (although technically it could work without the **kwargs option)
- add_causes(number, **kwargs)[source]#
Creates drug nodes related with each of the “Cancer” nodes already on the database
- Parameters
number (int) – From 0 to 9, the number under which the WikiData_IDs to process should ends. This allows us tho divide the work, although its not very elegant.
**kwargs – Any number of arbitrary keyword arguments
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Note
Here, there is no need to force c.WikiData_ID to not be null or “” because it will already be =
number(and, thus, exist)
- add_disease_info(number, **kwargs)[source]#
Adds info to “Disease” nodes for which its WikiData_ID ends in a given number. This way, only some of the nodes are targeted, and the Java Virtual Machine does not run out of memory
- Parameters
number (int) – From 0 to 9, the number under which the WikiData_IDs to process should ends. This allows us tho divide the work, although its not very elegant.
**kwargs – Any number of arbitrary keyword arguments
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Note
Here, there is no need to force c.WikiData_ID to not be null or “” because it will already be =
number(and, thus, exist)
- add_drug_external_ids(query='Wikidata_ID', **kwargs)[source]#
Adds some external IDs to any “Drug” nodes already present on the database. Since the PDB information had too much values which caused triple duplicates that overcharged the system, they were intentionally left out.
- Parameters
query (str) – One of [“DrugBank_ID”,”WikiData_ID”], a way to identify the nodes for which external IDs will be added.
**kwargs – Any number of arbitrary keyword arguments
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
We are forcing c.WikiData_ID to not be null or “”. This is not necessary if we are just building the wikidata database, because there will always be a WikiData_ID, but it is useful in the rest of the cases
- add_drugs(number, **kwargs)[source]#
Creates drug nodes related with each of the “Cancer” nodes already on the database
- Parameters
number (int) – From 0 to 9, the number under which the WikiData_IDs to process should ends. This allows us tho divide the work, although its not very elegant.
**kwargs – Any number of arbitrary keyword arguments
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Note
Here, there is no need to force c.WikiData_ID to not be null or “” because it will already be =
number(and, thus, exist)
- add_gene_info()[source]#
A Cypher Query that adds some external IDs and properties to “Gene” nodes already existing on the database. This query forces the genes to have a “found_in_taxon:homo_sapiens” label. This means that any non-human genes will not be annotated (.. TODO:: delete those)
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Note
Genomic Start and ends keep just the 2nd position, as reported in wikidata
Note
We are forcing c.WikiData_ID to not be null or “”. This is not necessary if we are just building the wikidata database, because there will always be a WikiData_ID, but it is useful in the rest of the cases
Todo
Might include P684 “Orthologues” for more info (it crashed java)
- add_genes(number, **kwargs)[source]#
Creates gene nodes related with each of the “Cancer” nodes already on the database
- Parameters
number (int) – From 0 to 9, the number under which the WikiData_IDs to process should ends. This allows us tho divide the work, although its not very elegant.
**kwargs – Any number of arbitrary keyword arguments
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Note
Here, there is no need to force c.WikiData_ID to not be null or “” because it will already be =
number(and, thus, exist)
- add_metabolite_info(query='ChEBI_ID', **kwargs)[source]#
A Cypher Query that adds some external IDs and properties to “Metabolite” nodes already existing on the database. Two kind of metabolites exist: those that are encoded by a given gene, and those that interact with a given drug. Both are adressed here, since they are similar, and, most likely, instances of proteins.
This function forces all metabolites to have a “found_in_taxon:human” target
The metabolites are not forced to be proteins, but if they are, this is kept in the “instance_of” record
- Parameters
query (str) – One of [“DrugBank_ID”,”WikiData_ID”], a way to identify the nodes for which external IDs will be added; default is “WikiData_ID”
**kwargs – Any number of arbitrary keyword arguments
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Todo
Might include P527 “has part or parts” for more info (it crashed java)
Note
We are forcing c.WikiData_ID to not be null or “”. This is not necessary if we are just building the wikidata database, because there will always be a WikiData_ID, but it is useful in the rest of the cases
- add_more_drug_info(query='WikiData_ID', **kwargs)[source]#
Creates some nodes that are related with each of the “Drug” nodes already existing on the database: routes of administration, targeted metabolites and approved drugs that tehy are been used in
- Parameters
query (str) – One of [“DrugBank_ID”,”WikiData_ID”], a way to identify the nodes for which external IDs will be added; default is “WikiData_ID”
**kwargs – Any number of arbitrary keyword arguments
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Todo
ADD ROLE to metabolite interactions
Note
This transaction has been separated in order to keep response times low
Note
We are forcing c.WikiData_ID to not be null or “”. This is not necessary if we are just building the wikidata database, because there will always be a WikiData_ID, but it is useful in the rest of the cases
- add_toomuch_metabolite_info()[source]#
A function that adds loads of info to existing “Metabolite” nodes. This was left out, first because it might be too much information, (specially when it is already availaible by clicking the “url” field), and because, due to it been so much, it crashes the JVM.
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
- add_wikidata_and_mesh_by_name()[source]#
A function that adds some MeSH nodes and WikiData_IDs to existing nodes, based on their Wikipedia Article Title.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- add_yet_more_drug_info(query='WikiData_ID', **kwargs)[source]#
Creates some nodes that are related with each of the “Drug” nodes already existing on the database: routes of administration, targeted metabolites and approved drugs that tehy are been used in
- Parameters
query (str) – One of [“DrugBank_ID”,”WikiData_ID”], a way to identify the nodes for which external IDs will be added; default is “WikiData_ID”
**kwargs – Any number of arbitrary keyword arguments
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
- find_instance_of_disease()[source]#
A Neo4J Cypher Statment that queries wikidata for instances of “Disease” nodes already present on the Database. Since these are expected to only affect humans, this subclasses should also, only affect humans
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Note
We are forcing c.WikiData_ID to not be null or “”. This is not necessary if we are just building the wikidata database, because there will always be a WikiData_ID, but it is useful in the rest of the cases
- find_subclass_of_disease()[source]#
A Neo4J Cypher Statment that queries wikidata for subclasses of “Disease” nodes already present on the Database. Since these are expected to only affect humans, this subclasses should also, only affect humans
- Returns
A CYPHER query that modifies the DB according to the CYPHER statement contained in the function.
- Return type
Note
We are forcing c.WikiData_ID to not be null or “”. This is not necessary if we are just building the wikidata database, because there will always be a WikiData_ID, but it is useful in the rest of the cases
- initial_cancer_discovery()[source]#
A Neo4J Cypher Statment that queries wikidata for Human Cancers. Since using the “afflicts:human” tag didnt have much use here, I used a simple workaround: Query wikidata for all humans, and, among them, find all of this for which their cause of death was a subclass of “Cancer” (Q12078). Unfortunaltely, some of them were diagnosed “Cancer” (Q12078), which is too general, so I removed it.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
CanGraph.QueryWikidata.main module#
A python module that leverages the functions present in the build_database
module to recreate selected parts of the the Wikidata database
using a graph format and Neo4J, and then provides an GraphML export file.
Please note that, to work, the functions here pre-suppose you have internet access, which will be used to access Wikidata’s SPAQL endpoint and write info to the Neo4J database
For more details on how to run this script, please consult the package’s README
